In response to the COVID-19 pandemic, the Microbial Analysis and Resource Services core and the Institute for Systems Genomics established a partnership in March 2020 to facilitate testing for SARS-Cov2, the virus that causes COVID-19. We have assembled a team among campus community partners to develop an integrated surveillance approach using existing expertise and adapting cutting edge molecular and data visualization technologies for our University. Since then, the team, led by Dr. Kendra Maas, has worked to develop and implement multiple testing workflows that have been deployed both locally at the University and across the state.
(last updated June 23, 2021)
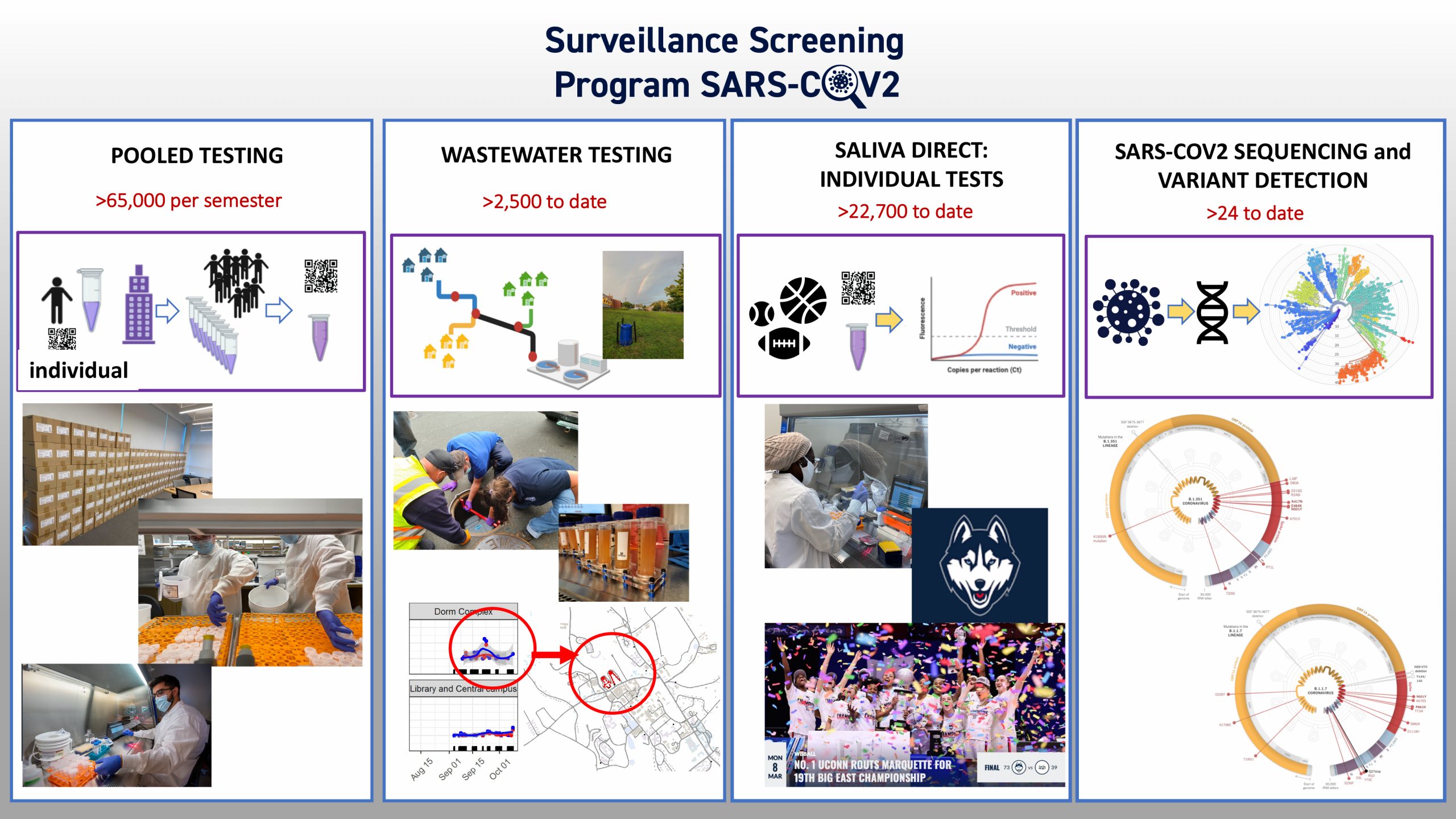
Pooled Gargle Testing
In support of student programs through Student Health and Wellness, and UConn Health, and external partners, our team developed a pooled testing strategy that implements a qRT-PCR test in gargle lavage samples. By pooling samples, we provide a broad surveillance approach that t is fast, affordable and scalable is critically needed to effectively screen members of the campus community for SARS-CoV2, support targeted responses to emerging clusters of cases, and contain and mitigate COVID-19, and thus prevent outbreaks. We are processing >65,000 samples per semester, representing a cumulative savings of $1.3-4.6M to the University.
Wastewater Testing
A composite sampler is placed in a manhole targeting a specific sewershed on campus. Daily collections from this pump are processed by concentrating viruses using nanoparticles and extracting RNA, followed by qRT-PCR. To date, 16 pumps have been placed on campus and samples are collected and tested daily (2,594 samples to date). Geographic Information Systems (GIS) mapping for UConn (developed by ITS and CLEAR) has allowed the team to identify the buildings producing the wastewater, thus delineating the presence of SARS-CoV2 in specific areas on campus (e.g. central wastewater processing plant, dormitory complexes, townships, etc).
Saliva Direct Testing
In support of the Division of Athletics, our team provides Saliva Direct tests daily, with >22,795 tests to date (June 23, 2021). These tests represent a cumulative savings of >$1.4M to the University, providing a rapid turnaround for test results and facilitating UConn’s participation in the national sports arena.
SARS-Cov2 Sequencing
Our team performs SARS-Cov2 sequencing for variant detection on multiple sample types and using multiple platforms. We process samples from Saliva Direct collections, nasal swabs, gargle lavage, and wastewater. Using ARTIC primers, we perform Illumina sequencing for high accuracy sequencing and Nanopore sequencing (ONT) for mobile sequencing applications. In addition, we have developed long-read sequencing applications of both RNA and DNA barcodes to span viral genome subregions among complex samples, such as wastewater. Our bioinformatics team is in the final stages of developing a pipeline for deconvolution and calling variants within complex mixtures of viral samples, as is found in wastewater collections.